Supplementary material for ``Reject Inference in Credit Scoring: a rationale review’’: tree biasedness
Adrien Ehrhardt
2021-01-10
Source:vignettes/appendix_tree_biasedness.Rmd
appendix_tree_biasedness.RmdData generation
m <- 10000
set.seed(21)
y <- rbinom(m, 1, 0.5)
data <- array(0, c(m, d + 1))
x <- array(0, c(m, d))
x[y == 0, ] <- mvrnorm(n = sum(y == 0), mu0, sigma0)
x[y == 1, ] <- mvrnorm(n = sum(y == 1), mu1, sigma1)
data <- as.matrix(cbind.data.frame(y = y, x = x))
rm(x)
rm(y)
train <- as.data.frame(data[1:(m / 2), ])
test <- as.data.frame(data[(m / 2 + 1):m, ])
train[, "y"] <- as.factor(train[, "y"])Loop over cut-off values
list_gini_arbre <- list()
list_gini_reglog <- list()
for (i in seq(0.2, 0.7, 0.05)) {
ind_refuses_arbre <- predict(modele_complet_arbre, train)[, 1] < i
ind_refuses_reglog <- predict(modele_complet_reglog, train, type = "response") < i
train_partiel_arbre <- train[!ind_refuses_arbre, ]
train_partiel_reglog <- train[!ind_refuses_reglog, ]
modele_partiel_arbre <- rpart(y ~ ., data = train_partiel_arbre, method = "class")
modele_partiel_reglog <- glm(y ~ ., data = train_partiel_reglog, family = binomial(link = "logit"))
list_gini_arbre <- append(list_gini_arbre, normalizedGini(test[, "y"], predict(modele_partiel_arbre, test)[, 2]))
list_gini_reglog <- append(list_gini_reglog, normalizedGini(test[, "y"], predict(modele_partiel_reglog, test, type = "response")))
}Figures
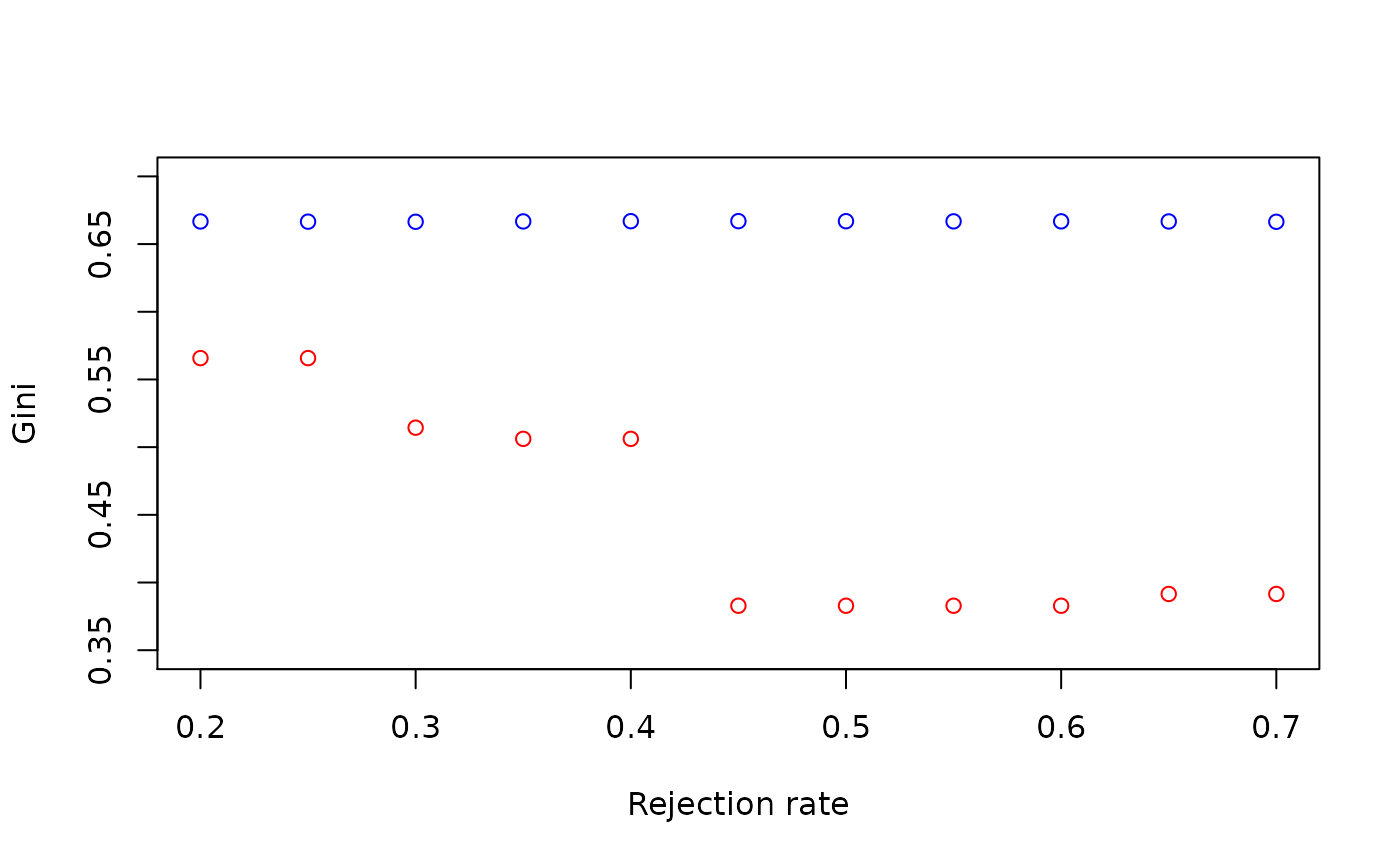
plot(seq(0.2, 0.7, 0.05),
list_gini_arbre,
ylim = c(0.35, 0.7),
xlab = "Rejection rate",
ylab = "Gini",
col = "red"
)
lines(seq(0.2, 0.7, 0.05), list_gini_reglog, ylim = c(0.35, 0.7), type = "p", col = "blue")
legend(1, 0.45,
pch = c(1, 1), lty = c(1, 1),
col = c("red", "blue"),
legend = c("Decision tree", "Logistic regression"),
cex = 1
)